Welcome To PhylomeDB6!
Your catalog of gene phylogenies.
What is phylomeDB?
PhylomeDB is a public database for complete catalogs of gene phylogenies (phylomes). It allows users to interactively explore the evolutionary history of genes through the visualization of phylogenetic trees and multiple sequence alignments. Moreover, phylomeDB provides genome-wide orthology and paralogy predictions which are based on the analysis of the phylogenetic trees. The automated pipeline used to reconstruct trees aims at providing a high-quality phylogenetic analysis of different genomes, including Maximum Likelihood tree inference, alignment trimming and evolutionary model testing. PhylomeDB includes also a public download section with the complete set of trees, alignments and orthology predictions. Finally, phylomeDB provides an advanced tree visualization interface based on the ETE toolkit, which integrates tree topologies, taxonomic information, domain mapping and alignment visualization in a single and interactive tree image.

ERGA PHYLOMES INICIATIVE
Call for collaborative projects proposal
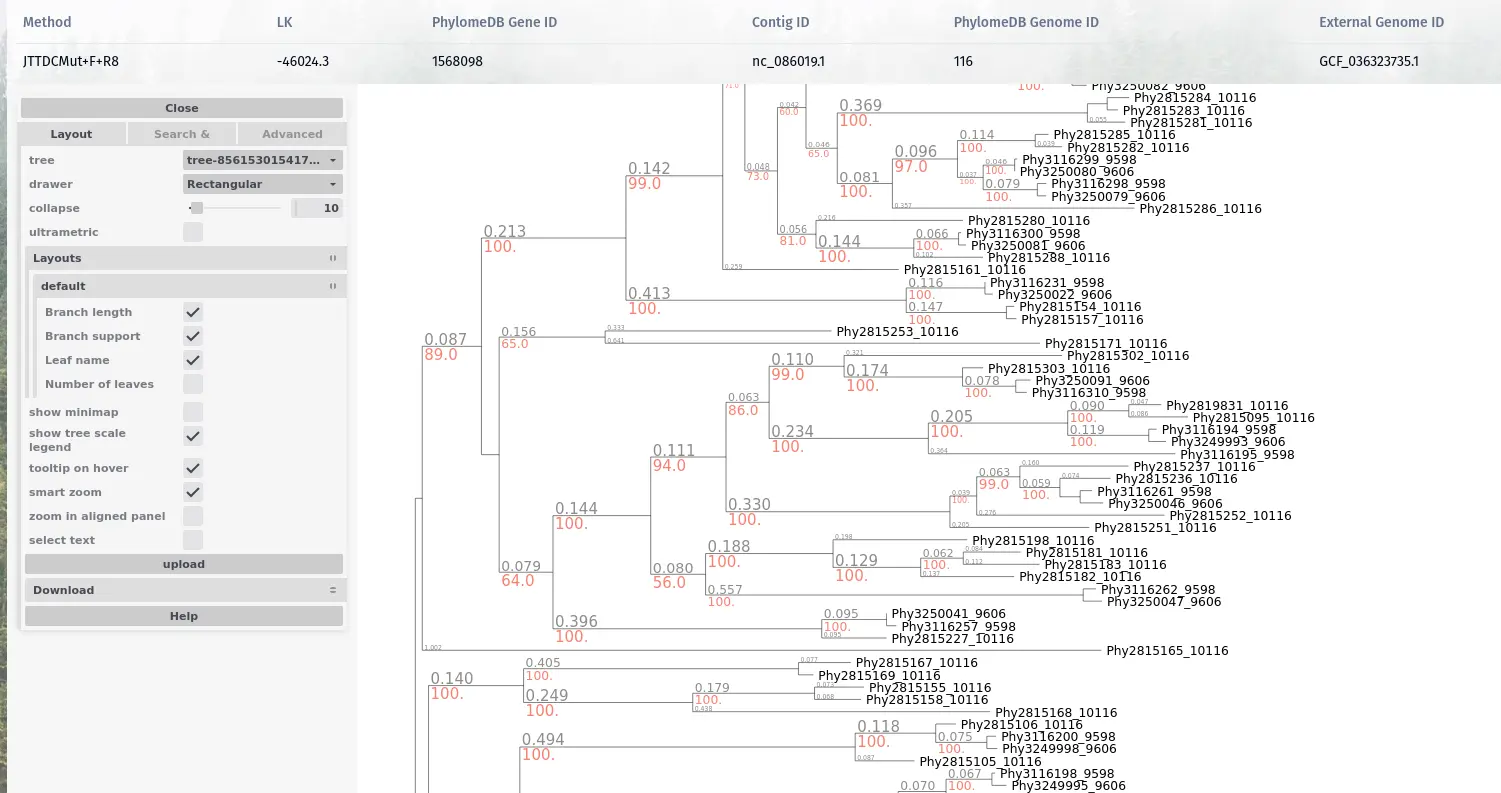
Phylogenetic trees
Representing the evolutionary relationships of homologous genes are the entry point for many evolutionary analyses.
Phylo Explorer
Decide bettter which phylome suits your need by browsing a set of species that the result phylomes must contain.

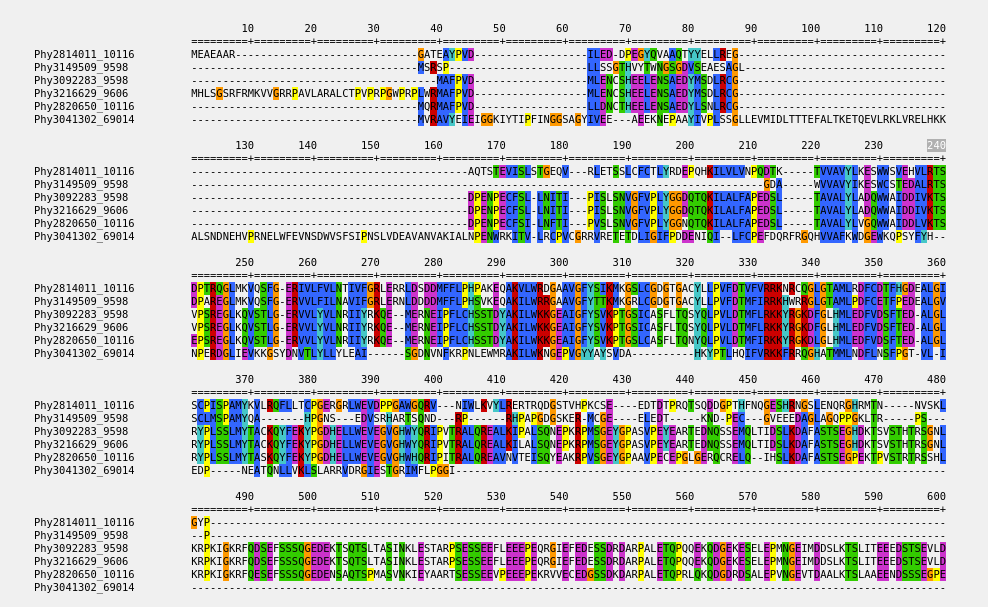
Alignments
PhylomeDB offers access to comprehensive multiple sequence alignments derived from phylogenetic analyses.
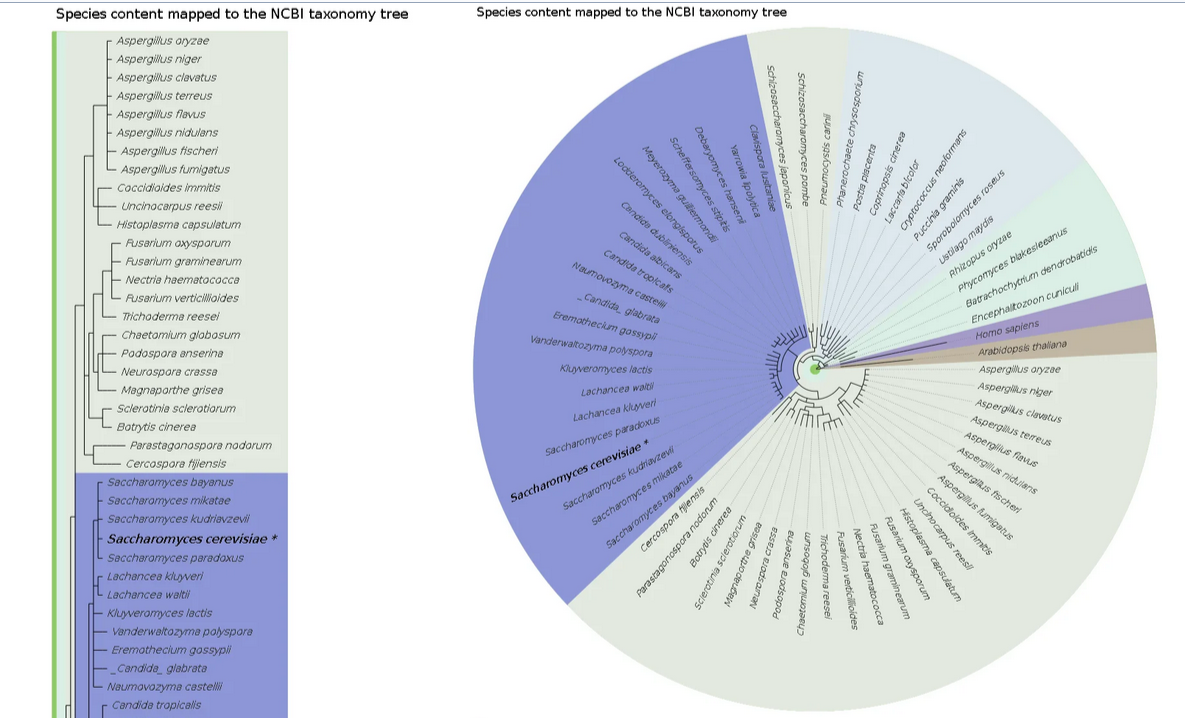
Species and taxonomy
PhylomeDB visually represents the relationships and classifications of various species within the NCBI taxonomy tree.